I have to do 3 things in here:
1. ADD PICTURES
2.
EDIT TEXT and improve overall legibility/readability and connect small
sentences into whole paragraphs to improve sense of the text. I can
choose to do this by any way I want but the information must be
retained. Its messy, clear it up
3. EXPAND IT
-
-
Bidirectional
not un-directional
-
Semi-conservative not conservative nor dispersive as shown in the
Meselson and Stahl experiment
and the script
Meselson and Stahl experiment
and the script
-
One origin in bacteria, multiple (thousands to ten of thousands of) origins in
eukaryotes
It takes about 8hours for DNA to replicate in eukaryotes. 50bp/s in eukaryotes and 1000-2000bp/s in prokaryotes its speed. My lecture notes though differ with this statement, that 1.5hrs in yeast and 24hrs in eukaryotes is needed to replicate the DNA.
It takes about 8hours for DNA to replicate in eukaryotes. 50bp/s in eukaryotes and 1000-2000bp/s in prokaryotes its speed. My lecture notes though differ with this statement, that 1.5hrs in yeast and 24hrs in eukaryotes is needed to replicate the DNA.
-
DNA pol: always polymerize/synthesize in a 5’->3’
direction, adds nucleotides in order. Forms phosphodiester bond, catalyses the
addition of nucleotide to the 3’ OH of the last nucleotide of the growing
strand
Ori C (the origin of replication in E. Coli) is 13 base pair sequence rich in AT pairs repeated 5 times.
Ori C (the origin of replication in E. Coli) is 13 base pair sequence rich in AT pairs repeated 5 times.
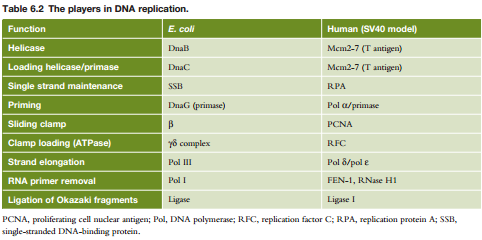
In E. coli:
I …
in repair and replication
II
DNA repair
III
principal DNA replication enzyme
IV
DNA repair
V DNA repair
-
Proofreading
-
Sliding
clamp
-
Unwinding the helix: topoisomerases, helicase,
single strand binding proteins ssbp
-
First initiator proteins in e coli bind to AT
rich origin, forms a spiral weakening the double helix and allows helicase and
other replication enzymes to attach
-
Priming; dna pol can only extend dna strand at
the 3’ end; requires OH to add to. Each new strand starts with a short primer
of 10-30 RNA bases complementary to DNA. Primer is formed by an enzyme call
primase. Primase does not
require an existing 3′ OH for synthesis, unlike DNA polymerase. RNA
Primer is later removed by DNA polymerase I and replaced with correct DNA
sequence
-
DNA Polymerase can add to RNA primer: Polymerase
switching occurs. Primase finishes synthesising the primer. The clamp attaches
to the end of the primer. DNA polymerase attaches to the clamp
-
Semi-discontinuous; leading and lagging strand (leading
-> continuous: new strand is made without breaks although there are
fragments produced on the leading strand because of repair; lagging -> discontinuous: new strand is
made in short fragments/okazaki fragments)
-
Lagging
strand in bacteria: Primase synthesizes RNA primer, DNA pol III continuous
to synthesize full okazaki fragments and stops when it reaches next primer. Pol
III is replaced by Pol I that hydrolyses RNA primer and replaces RNA primer
with DNA and leaves a ‘nick’, an unmade phosphodiester bond between
discontinuous okazaki fragments. DNA ligase seals the nick by making bond
-
Lagging
strand in eukaryotes: Primase
synthesises RNA primer and DNA pol δ
continuous to synthesise full okazaki fragment and continuous until it reaches
primer then displaces the primer leaving a flap. Flap endonuclease Fen1 cleaves
the flap DNA, leaves a nick between discontinuous okazaki fragments that DNA
ligase seals the nick by making a bond.
-
DNA polymerase moves in different directions on
the leading and lagging strands but overall, polymerase on both strands moves
with the replication fork because the lagging strand makes a loop around. Joint
regulation of lead and lagging strand synthesis
-
Bacterial
replisome: A molecular machine for DNA replication. Contains 2 copies of
DNA Polymerase III (Lead & lagging strand). E. coli can replicate
its whole genome of 5 million base pairs in 40 min. With 2 replication forks,
each replisome works at 1,000 nucleotides/sec. Combines speed with accuracy
-
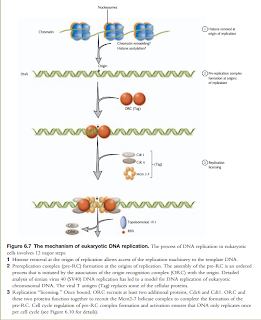
Eukaryotic
Replication Fork: Eukaryotic replication fork is similar to
replisome. Leading and lagging strand synthesis is coupled but not one unit. DNA
polymerase δ replicates the leading strand. DNA polymerase ε the lagging strand
-
Like a
trombone? Proposed by
Alberts et al.In the trombone model, DNA on the lagging strand is looped
around, so that the polymerases at the fork are working in the same direction. Every
so often, the lagging strand DNA would be released and re-oriented at the fork
-
Chromatin
in Eukaryotic replication Chromatin
is the reason replication in eukaryotes is slower. Eukaryotic linear
chromosomes of chromatin. Nucleosomes –are disrupted by the replication fork,
then re-bind after the fork has passed – why replication. Half of the parental H3 and H4 go to one
daughter, half to the other. Called parental histone segregation. ASF1 guides
parental H3 and H4 histones to their new position. New histones delivered with
chromatin assembly factor 1 (CAF-1) to replication fork. The correct
recruitment of histones, and maintenance of chemical modifications such as
methylation underpin epigenetic inheritance
-
Prokaryotic
replication: Termination In
bacterial circular chromosomes, termination of replication occurs at the ter
site. Ter is on the opposite side of the chromosome from ori. ter is bound by Tus – a replicator
terminator protein. When replication
forks hit Tus they stop, and disassembly of the replication complex occurs
-
Eukaryotic
replication: Termination Replication
can continue to the end of a chromosome except that the lagging strand requires
enzymes (primase) to bind ahead of the replication fork. Therefore the lagging
strand will get shorter with every replication. Telomeres are sacrificial ends
of chromosomes to protect coding DNA. Telomerase contains RNA and adds
telomeric DNA sections onto the 3’ end
-
Chromosome
Ends – Telomeres protect chromosome ends. Chromosomes get shorter. But only
telomere sequence is lost…for a while. Telomeres range in length from ~100bp to > 20,000bp, depending on
species. Telomerase contains RNA that acts as a template for the
extension of the DNA with hexamer (TTAGGG) repeats. These provide a site for
primer synthesis in replication. Excess hexamer repeats can then be removed. Telomerase extends chromosomes although
most normal cells do not express the telomerase and so lose telomeres with each
division. In humans telomerase is active in germ cells, epidermal skin cells,
follicular hair cells, most cancer cells, some stem cells probably and in vitro
immortalised cells.
S Some pictures good pictures:
S Some pictures good pictures:












No comments:
Post a Comment